We want to test out this nf-core workflow: https://github.com/nf-core/differentialabundance for our RNAseq data output.
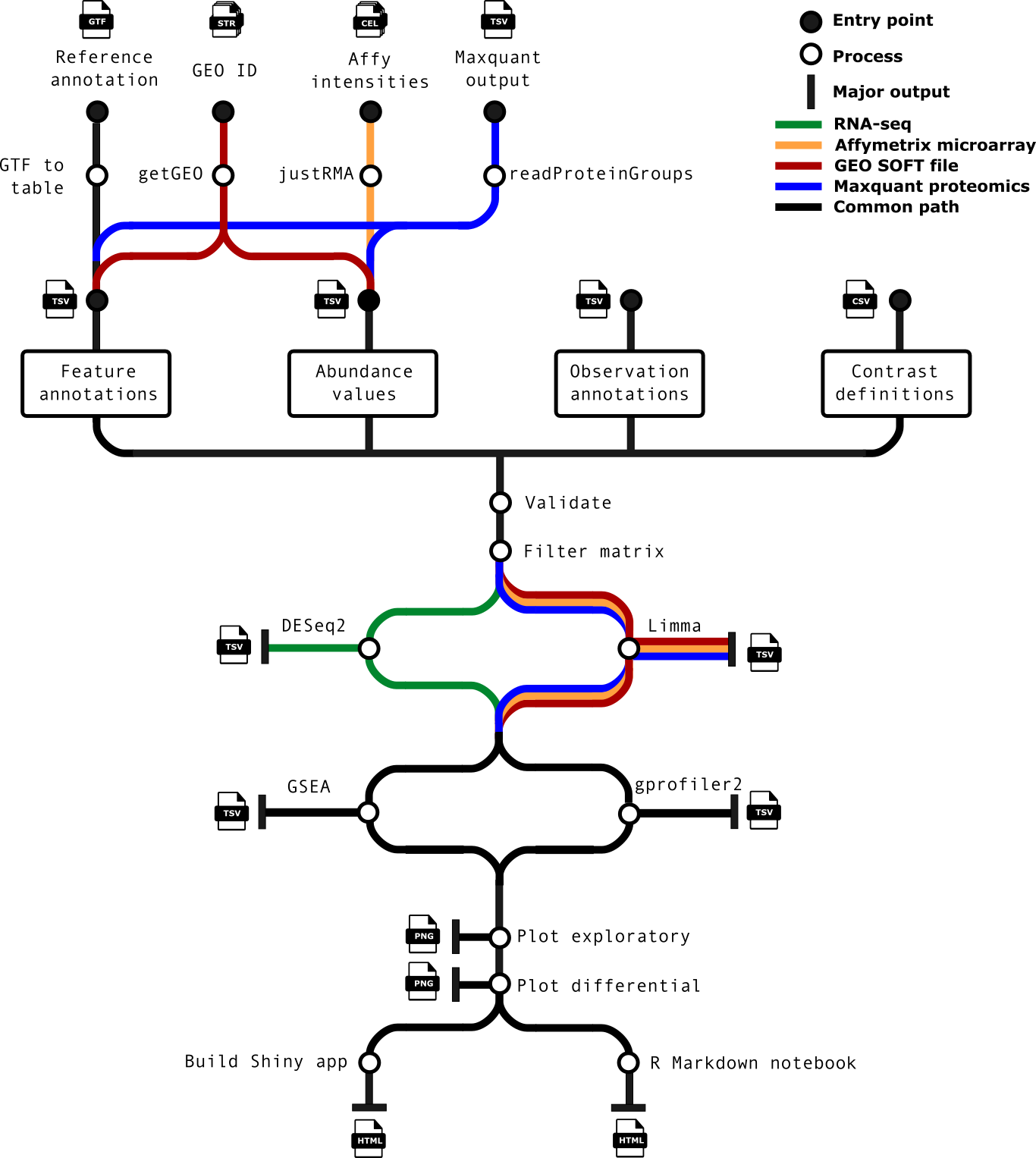
Required files
Observations / Samplesheet input: --input rnaseq_difabundance_fullset_samplesheet.csv
sample,fastq_1,fastq_2,condition
SRX13037862,/gscratch/scrubbed/strigg/analyses/20241205_FetchNGS/fastq/SRX13037862_SRR16844801.fastq.gz,,tolerant
SRX13037863,/gscratch/scrubbed/strigg/analyses/20241205_FetchNGS/fastq/SRX13037863_SRR16844800.fastq.gz,,tolerant
SRX18040005,/gscratch/scrubbed/strigg/analyses/20241205_FetchNGS/fastq/SRX18040005_SRR22059186_1.fastq.gz,/gscratch/scrubbed/strigg/analyses/20241205_FetchNGS/fastq/SRX18040005_SRR22059186_2.fastq.gz,tolerant
SRX18040006,/gscratch/scrubbed/strigg/analyses/20241205_FetchNGS/fastq/SRX18040006_SRR22059167_1.fastq.gz,/gscratch/scrubbed/strigg/analyses/20241205_FetchNGS/fastq/SRX18040006_SRR22059167_2.fastq.gz,tolerant
RNAseq data: I used the salmon.merged.gene_length_scaled.tsv files and merged them in R to be merged_gene_counts_scaled.tsv.
--matrix 'merged_gene_counts_scaled.tsv'
Contrasts information: --contrasts rnaseq_diffabundance_contrasts.csv
id,variable,reference,target
condition_sensitive_tolerant,condition,sensitive,tolerant
Feature annotations --gtf '[path to gtf file]'
/gscratch/srlab/elstrand/genomes/C_virginica/mod_GCF_002022765.2_C_virginica-3.0_genomic.gtf.gz
Transfer files
scp C:\Users\EmmaStrand\MyProjects\Resilience_Biomarkers_Aquaculture\Cvirg_Pmarinus_RNAseq\data\rnaseq_gene_counts\merged_gene_counts_scaled.tsv elstrand@klone.hyak.uw.edu:/mmfs1/gscratch/scrubbed/elstrand/Cvir_disease_meta/differential_abundance/merged_gene_counts_scaled.tsv
scp C:\Users\EmmaStrand\MyProjects\Resilience_Biomarkers_Aquaculture\Cvirg_Pmarinus_RNAseq\data\differential_abundance_sheets\rnaseq_difabundance_fullset_samplesheet.csv elstrand@klone.hyak.uw.edu:/mmfs1/gscratch/scrubbed/elstrand/Cvir_disease_meta/differential_abundance/rnaseq_difabundance_fullset_samplesheet.csv
scp C:\Users\EmmaStrand\MyProjects\Resilience_Biomarkers_Aquaculture\Cvirg_Pmarinus_RNAseq\data\differential_abundance_sheets\rnaseq_diffabundance_contrasts.csv elstrand@klone.hyak.uw.edu:/mmfs1/gscratch/scrubbed/elstrand/Cvir_disease_meta/differential_abundance/rnaseq_diffabundance_contrasts.csv
Downloading Shinyngs to the conda environment I created for nextflow
The output of differential abundance uses r-shinyngs so I’m downloading that within the mamba environment we I created called nextflow.
## Activate current environment
mamba activate nextflow
## Install r-shinyngs
conda config --add channels bioconda
mamba install r-shinyngs
Script
Trying to run this on the full dataset.
differential_abundance_full.sh
#!/bin/bash
#SBATCH --account=srlab
#SBATCH --error="%x_error.%j" #if your job fails, the error report will be put in this file
#SBATCH --output="%x_output.%j" #once your job is completed, any final job report comments will be put in this file
#SBATCH --partition=cpu-g2-mem2x
#SBATCH --nodes=1
#SBATCH --time=1-20:00:00
#SBATCH --mem=50G
#SBATCH --ntasks=7
#SBATCH --cpus-per-task=2
## Set paths
samplesheet="/gscratch/scrubbed/elstrand/Cvir_disease_meta/differential_abundance/rnaseq_difabundance_fullset_samplesheet.csv"
contrasts="/gscratch/scrubbed/elstrand/Cvir_disease_meta/differential_abundance/rnaseq_diffabundance_contrasts.csv"
gene_counts="/mmfs1/gscratch/scrubbed/elstrand/Cvir_disease_meta/differential_abundance/merged_gene_counts_scaled.tsv"
output_dir="/mmfs1/gscratch/scrubbed/elstrand/Cvir_disease_meta/differential_abundance/results"
gtf="/gscratch/srlab/elstrand/genomes/C_virginica/mod_GCF_002022765.2_C_virginica-3.0_genomic.gtf"
## Run differential abundance workflow
nextflow run nf-core/differentialabundance -resume \
-profile rnaseq,singularity \
--input ${samplesheet} \
--contrasts ${contrasts} \
--matrix ${gene_counts} \
--outdir ${output_dir} \
--gtf ${gtf} \
--features_gtf_feature_type exon
To run:
- Activate conda environment:
mamba activate nextflow - Run script:
sbatch differential_abundance_full.sh
Notes:
- 2-27-2025 Tried to run on full dataset. Got error from gtf file. There is something about the file that it can’t read.. Possible that it’s looking for ‘transcript’ and can’t find any? This R script is within the workflow so if I wanted to change any parameters then I’d have to include a custom config file.
ERROR ~ Error executing process > 'NFCORE_DIFFERENTIALABUNDANCE:DIFFERENTIALABUNDANCE:GTF_TO_TABLE (mod_GCF_002022765)'
Caused by:
Process `NFCORE_DIFFERENTIALABUNDANCE:DIFFERENTIALABUNDANCE:GTF_TO_TABLE (mod_GCF_002022765)` terminated with an error exit status (1)
Command executed:
gtf2featureAnnotation.R \
--gtf-file mod_GCF_002022765.2_C_virginica-3.0_genomic.gtf \
--output-file mod_GCF_002022765.anno.tsv \
\
--feature-type 'transcript' --first-field 'gene_id'
cat <<-END_VERSIONS > versions.yml
"NFCORE_DIFFERENTIALABUNDANCE:DIFFERENTIALABUNDANCE:GTF_TO_TABLE":
atlas-gene-annotation-manipulation: 1.1.1
END_VERSIONS
Command exit status:
1
Command output:
[1] "Reading mod_GCF_002022765.2_C_virginica-3.0_genomic.gtf elements of type transcript"
I tried awk '$3 == "transcript"' mod_GCF_002022765.2_C_virginica-3.0_genomic.gtf to see if there are any transcripts and this returned nothing. I then tried awk '{print $3}' mod_GCF_002022765.2_C_virginica-3.0_genomic.gtf | sort | uniq and this resulted in:
CDS Crassostrea exon gene start_codon stop_codon
The gtf file I’m using looks like:
#gtf-version 2.2
#!genome-build C_virginica-3.0
#!genome-build-accession NCBI_Assembly:GCF_002022765.2
#!annotation-source NCBI Crassostrea virginica Annotation Release 100
NC_035780.1 Gnomon gene 13578 14594 . + . gene_id "LOC111116054"; db_xref "GeneID:111116054"; gbkey "Gene"; gene "LOC111116054"; gene_biotype "lncRNA";
NC_035780.1 Gnomon exon 13578 13603 . + . gene_id "LOC111116054"; transcript_id "XR_002636969.1"; db_xref "GeneID:111116054"; gbkey "ncRNA"; gene "LOC111116054"; model_evidence "Supporting evidence includes similarity to: 100% coverage of the annotated genomic feature by RNAseq alignments, including 1 sample with support for all annotated introns"; product "uncharacterized LOC111116054"; exon_number "1";
NC_035780.1 Gnomon exon 14237 14290 . + . gene_id "LOC111116054"; transcript_id "XR_002636969.1"; db_xref "GeneID:111116054"; gbkey "ncRNA"; gene "LOC111116054"; model_evidence "Supporting evidence includes similarity to: 100% coverage of the annotated genomic feature by RNAseq alignments, including 1 sample with support for all annotated introns"; product "uncharacterized LOC111116054"; exon_number "2";
NC_035780.1 Gnomon exon 14557 14594 . + . gene_id "LOC111116054"; transcript_id "XR_002636969.1"; db_xref "GeneID:111116054"; gbkey "ncRNA"; gene "LOC111116054"; model_evidence "Supporting evidence includes similarity to: 100% coverage of the annotated genomic feature by RNAseq alignments, including 1 sample with support for all annotated introns"; product "uncharacterized LOC111116054"; exon_number "3";
NC_035780.1 Gnomon gene 28961 33324 . + . gene_id "LOC111126949"; db_xref "GeneID:111126949"; gbkey "Gene"; gene "LOC111126949"; gene_biotype "protein_coding";
NC_035780.1 Gnomon exon 28961 29073 . + . gene_id "LOC111126949"; transcript_id "XM_022471938.1"; db_xref "GeneID:111126949"; gbkey "mRNA"; gene "LOC111126949"; model_evidence "Supporting evidence includes similarity to: 3 Proteins, and 100% coverage of the annotated genomic feature by RNAseq alignments, including 21 samples with support for all annotated introns"; product "UNC5C-like protein"; exon_number "1";
3-10-2025: I figured out that I can change the feature type that the workflow is looking for. Originally this was ‘transcript’, but I’ll try to change it to ‘exon’ and see if that works. This is adding the --features_gtf_feature_type flag to the workflow.
3-12-2025: I had gtf.gz" at the end of the gtf file. I removed .gz and tried again. Nextflow couldn’t find the file bc of the extension. Running again. I also changed the path of output files to be within differential_abundance/scripts. This is running! Check back in later.
I ran into this issue with the gff file. This is saying that line 1531480 has 8 columns which might be different than the rest? We have had many issues with this gtf file that I’m wondering if this workflow is worth it.
[1] "Reading mod_GCF_002022765.2_C_virginica-3.0_genomic.gtf elements of type exon"
Error in readGFF(filepath, version = version, filter = filter) :
reading GFF file: line 1531480 has less than 8 tab-separated columns
Calls: import ... import -> import -> .local -> readGFFAsGRanges -> readGFF
3-13-2025: I’m trying this workflow on Cgigas to see if I can get it work with a different gtf file. This Cgigas group was determined to be the tolerant group so there isn’t a comparison. I made 2 of those samples ‘test’ and 3 of those samples ‘experiment’ just to get the pipeline going.
Downloading genomes and rnaseq data again
wget https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/963/853/765/GCF_963853765.1_xbMagGiga1.1/GCF_963853765.1_xbMagGiga1.1_genomic.fna.gz
wget https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/963/853/765/GCF_963853765.1_xbMagGiga1.1/GCF_963853765.1_xbMagGiga1.1_genomic.gff.gz
wget https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/963/853/765/GCF_963853765.1_xbMagGiga1.1/GCF_963853765.1_xbMagGiga1.1_genomic.gtf.gz
wget https://gannet.fish.washington.edu/emma.strand/rnaseq/Cvir_Prkns_rnaseq_dataset3_Cgigas/salmon.merged.gene_counts_length_scaled.tsv
scp C:\Users\EmmaStrand\MyProjects\Resilience_Biomarkers_Aquaculture\Cvirg_Pmarinus_RNAseq\data\differential_abundance_sheets\rnaseq_difabundance_dataset3_Cgigas_samplesheet.csv elstrand@klone.hyak.uw.edu:/mmfs1/gscratch/scrubbed/elstrand/Cgigas_disease_meta/samplesheets/rnaseq_difabundance_dataset3_Cgigas_samplesheet.csv
scp C:\Users\EmmaStrand\MyProjects\Resilience_Biomarkers_Aquaculture\Cvirg_Pmarinus_RNAseq\data\differential_abundance_sheets\rnaseq_diffabundance_contrasts_Cgigas_test.csv elstrand@klone.hyak.uw.edu:/mmfs1/gscratch/scrubbed/elstrand/Cgigas_disease_meta/samplesheets/rnaseq_diffabundance_contrasts_Cgigas_test.csv
differential_abundance_Cgigas.sh
#!/bin/bash
#SBATCH --account=srlab
#SBATCH --error="%x_error.%j" #if your job fails, the error report will be put in this file
#SBATCH --output="%x_output.%j" #once your job is completed, any final job report comments will be put in this file
#SBATCH --partition=cpu-g2-mem2x
#SBATCH --nodes=1
#SBATCH --time=1-20:00:00
#SBATCH --mem=50G
#SBATCH --ntasks=7
#SBATCH --cpus-per-task=2
## Set paths
samplesheet="/gscratch/scrubbed/elstrand/Cgigas_disease_meta/samplesheets/rnaseq_difabundance_dataset3_Cgigas_samplesheet.csv"
contrasts="/gscratch/scrubbed/elstrand/Cgigas_disease_meta/samplesheets/rnaseq_diffabundance_contrasts_Cgigas_test.csv"
gene_counts="/gscratch/scrubbed/elstrand/Cgigas_disease_meta/dataset3_Cgigas/salmon.merged.gene_counts_length_scaled.tsv"
output_dir="/gscratch/scrubbed/elstrand/Cgigas_disease_meta/differential_abundance_results"
gtf="/gscratch/srlab/elstrand/genomes/C_gigas/GCF_963853765.1_xbMagGiga1.1_genomic.gtf.gz"
## Run differential abundance workflow
nextflow run nf-core/differentialabundance -resume \
-profile rnaseq,singularity \
--input ${samplesheet} \
--contrasts ${contrasts} \
--matrix ${gene_counts} \
--outdir ${output_dir} \
--gtf ${gtf}
To run:
- Activate conda environment:
mamba activate nextflow - Run script:
sbatch differential_abundance_Cgigas.sh
This worked!!! Bummer that the gtf file for Cvirginica was giving errors.
Pushing output to gannet:
cd /gscratch/scrubbed/elstrand/Cgigas_disease_meta/differential_abundance_results
rsync --archive --verbose --progress * emma.strand@gannet.fish.washington.edu:/volume2/web/emma.strand/differentialabundance/Cgigas_test
Examples from other projects
Riss ran this on thier rnaseq from sea urchins. They ended up with log2change values that seemed unrealistic so they ended up doing this on their own. I wonder if this was because the output from rnaseq was run with –deseq_vst so if the differential abundance workflow is also normalizing these value it would be a problem..?
#!/bin/bash
#SBATCH --error=output_messages/"%x_error.%j" #if job fails, error report is put into this file
#SBATCH --output=output_messages/"%x_output.%j" #final job report
#SBATCH --partition=short
#SBATCH --nodes=1
#SBATCH --time=24:00:00
#SBATCH --job-name=nfcore_differentialabundance_controlsovertime_RMK
#SBATCH --mem=50GB
#SBATCH --ntasks=24
#SBATCH --cpus-per-task=2
#Change directory to where data files are
cd /work/gmgi/Urchin/nfcoreRNAseq
#Load the 'singularity' container version 3.10.3
module load singularity/3.10.3
#Load the nextflow module, most recent version on Discovery
module load nextflow/24.04.4
#####NEXTFLOW DifferentialAbundance Pipeline w RNAseq Output#########
nextflow -log ./nextflow.log run nf-core/differentialabundance \
--max_cpus 24 \
--input /work/gmgi/Urchin/nfcoreRNAseq/controlsonly_samplesheet.csv \
--contrasts /work/gmgi/Urchin/nfcoreRNAseq/controlsonly_timecontrasts.csv \
--matrix /work/gmgi/Urchin/nfcoreRNAseq/nfcoreOutput/star_salmon/salmon.merged.gene_counts.tsv \
--transcript_length_matrix /work/gmgi/Urchin/nfcoreRNAseq/nfcoreOutput/star_salmon/salmon.merged.gene_lengths.tsv \
--outdir /work/gmgi/Urchin/nfcoreRNAseq/nfcoreDiffAbund_ControlsOverTimeOutput \
--gtf /work/gmgi/Urchin/GenomesAndAnnotationFiles/Spur_5.0.60.gtf.gz \
-profile rnaseq,singularity