This is a meta analysis of 4 RNAseq datasets each comparing different oysters infected with Perkinsus marinus.
I first got all the meta data from the SRA Run Selector (e.g. https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRP301630) I downloaded this csv file by clicking the ‘Metadata’ button under the download heading.
I then got lists of accessions and concatenated them into one list (ids.csv) to be fed to the FetchNGS pipeine.
Then I ran FetchNGS to get all the datasets
FetchNGS
# create a screen session
screen -S nextflow
# request a compute node (mem and time requests can be modified)
salloc -A srlab -p cpu-g2-mem2x -N 1 -c 1 --mem=16GB --time=12:00:00
# load the nextflow environment
mamba activate nextflow
# run nextflow pipeline
nextflow run \
nf-core/fetchngs \
-resume \
-c /gscratch/srlab/strigg/bin/uw_hyak_srlab.config \
--input /gscratch/scrubbed/strigg/analyses/20241104_RNAseq/ids.csv \
--outdir /gscratch/scrubbed/strigg/analyses/20241104_RNAseq \
--download_method sratools
11/08/2024
I ended up canceling the job (CTRL + c) because it was taking too long to run. It seemed stalled out:
- nextflow.log file
- trace file
- timeline report
- summary report: note it says completed even though I did CTRL + c
- I exited the screen session as follows
screen -XS nextflow quit
I reran the job in a new screen session and new directory (20241108_FetchNGS).
# create a screen session
screen -S nextflow
# request a compute node (mem and time requests can be modified)
salloc -A srlab -p cpu-g2-mem2x -N 1 -c 1 --mem=16GB --time=12:00:00
# load the nextflow environment
mamba activate nextflow
# run nextflow pipeline
nextflow run \
nf-core/fetchngs \
-resume \
-c /gscratch/srlab/strigg/bin/uw_hyak_srlab.config \
--input /gscratch/scrubbed/strigg/analyses/20241104_RNAseq/ids.csv \
--outdir /gscratch/scrubbed/strigg/analyses/20241108_RNAseq \
--download_method sratools
It completed in 1.5 hours which is crazy fast. 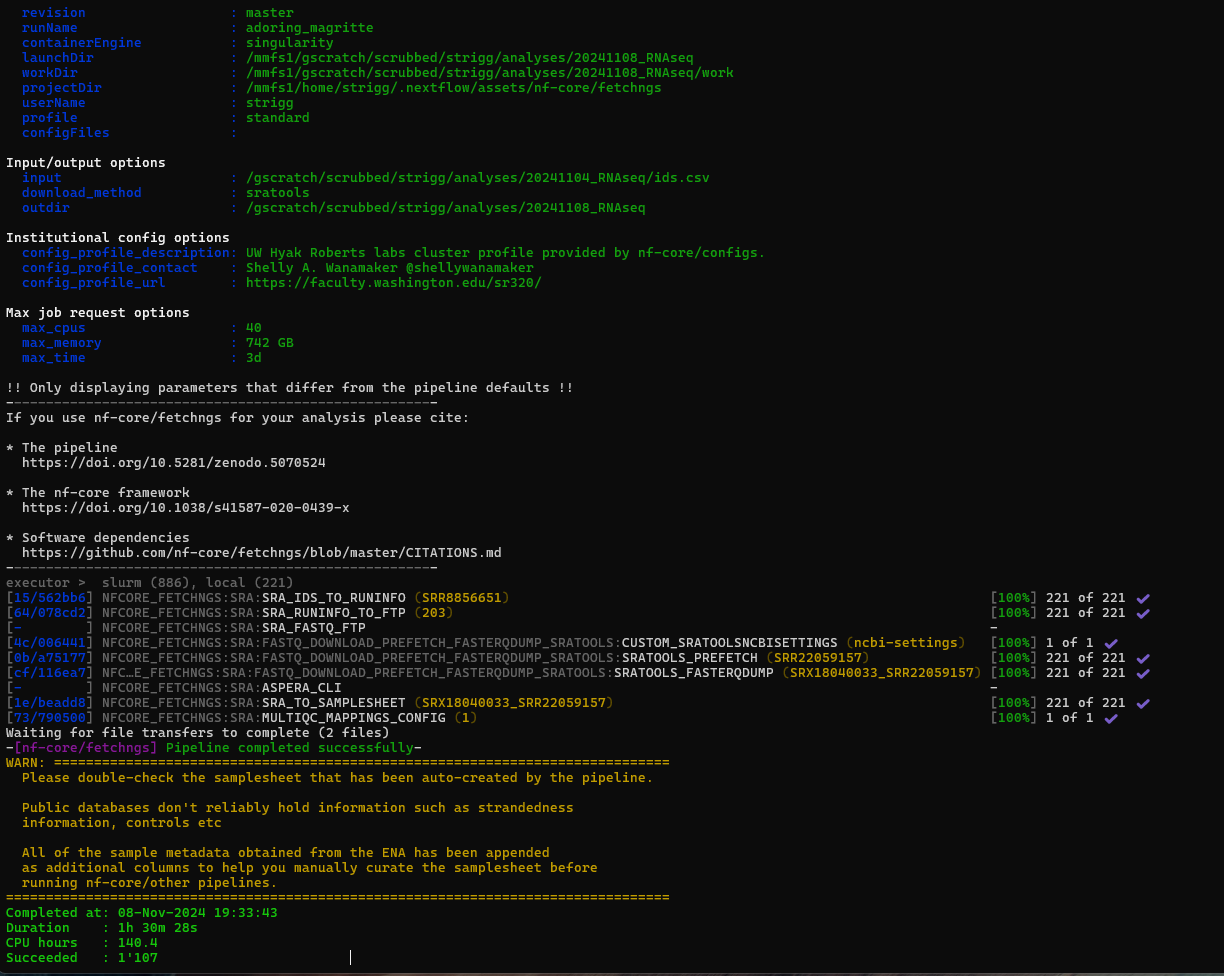 .
.
Summary Report here: https://gannet.fish.washington.edu/metacarcinus/USDA_MetaOmics/Cvirg_RNAseq/20241108_FetchNGS/pipeline_info/execution_report_2024-11-08_18-03-14.html
Timeline Report here: https://gannet.fish.washington.edu/metacarcinus/USDA_MetaOmics/Cvirg_RNAseq/20241108_FetchNGS/pipeline_info/execution_timeline_2024-11-08_18-03-14.html
Log file here: https://gannet.fish.washington.edu/metacarcinus/USDA_MetaOmics/Cvirg_RNAseq/20241108_FetchNGS/nextflow.log
Samplesheet is here: https://gannet.fish.washington.edu/metacarcinus/USDA_MetaOmics/Cvirg_RNAseq/20241108_FetchNGS/samplesheet/samplesheet.csv
genome for C. virginica
https://www.ncbi.nlm.nih.gov/datasets/genome/?taxon=6565
Next steps
- Run fastqc on reads to see if any trimming needs to be done
- Run Emma’s RNAseq pipeline